(added mention of Pull Mode) |
LociOiling (talk | contribs) (Freshen up a bit, add new images, add category.) |
||
| (6 intermediate revisions by 6 users not shown) | |||
| Line 1: | Line 1: | ||
| + | [[File:Helix_Loop_Sheet.png|thumb|400px|Sections of [[Loop|loop]], circled in blue, connect a [[Helix|helix]] and a [[Sheet|sheet]], shown in dashed boxes. Blue-and-white spirals show [[Hydrogen bond|hydrogen bonds]]. The loop at the top has seven [[Segment|segments]], making a relatively long loop.]] |
||
| − | The loop is one of three basic structures in FoldIt. It appears as a narrow cylindrical cord of arbitrary length, with kinks and [[sidechain|sidedchains]] corresponding to the constituent [[amino acids]]. In a sense, it is the most basic structure, representing any kind of structure that does not conform well enough to the [[helix]] or [[sheet]] forms. It is highly flexible, being easily reshaped by use of the various [[tools]] in [[Pull Mode]]. It is possible for loops to hydrogen bond with nearby structures, including adjacent amino acids, but this will not be visible unless the area of interest is turned into sheets. |
||
| + | [[Loop]] is one of three types of [[Secondary Structure|secondary structure]] seen in Foldit. Unlike [[Helix|helix]] and [[Sheet|sheet]], loop doesn't have a predictable shape. |
||
| + | |||
| + | In Foldit, using one of the "cartoon" [[View Options|view options]], sections of loop appear as a thin tube. Loop doesn't have the heavy tube and spiral shape of a helix, or the flat zigzag shape of sheets. |
||
| + | |||
| + | [[File:Long_loop.png|thumb|400px|A 12-segment loop connects two helixes. This pattern is found in the natural protein [https://www.rcsb.org/structure/2ftk 2FTK], the basis of "Rosetta Decoy 10", seen periodically as a [[Revisiting puzzle|revisiting puzzle]]. [[Ideal Loops|Ideal loops]] in Foldit are normally much shorter, but real proteins may contain long sections of loop.]] |
||
| + | Loop doesn't form the regular patterns of [[Hydrogen bond|hydrogen bonds]] between [[Protein backbone|backbone]] atoms that define helix and sheet. The lack of a regular pattern of bonds helps make loop sections more flexible than sheet or helix. In Foldit terms, however, any unbonded atoms represent a missed scoring opportunity, so there's an incentive to keep loop sections short, at least in [[Design puzzle|design puzzles]]. |
||
| + | |||
| + | In real proteins, there are often long sections of loop that meander between helixes and sheets. |
||
| + | |||
| + | In design puzzles, Foldit scientists have identified loop sections as a problem area. See [[Ideal Loops]] for discussion of the new features added to the game to address this issue. With more ideal loops, players are able to design proteins that actually fold up on their own. See [[Design puzzle results|design puzzle results]] for the latest on successful player designs. |
||
| + | |||
| + | The [[Blueprint]] tool contains predefined shapes that can be used as a starting point for designing ideal loops. The loop portions of these shapes are short -- only two to five segments. It's possible to design longer loops that satisfy the "ideal loops" condition found in many design puzzles, but it's not necessarily easy. |
||
| + | |||
| + | ==The Purpose of Loops== |
||
| + | Helixes and sheets tend to stabilize the [[Core|core]] of the protein. Loops are more likely to be found near the [[Surface|surface]] of the protein. Not surprisingly, loops tend to be rich in [[Hydrophilic|hydrophilic]] sidechains. The hydrophilics in loops make hydrogen bonds with the surrounding water more than with adjacent amino acids, helping make loops more flexible than helices and sheets. |
||
| + | |||
| + | Far from just being “connectors,” loops are often involved in the function of the protein, such as being part of the active site of an [[wikipedia:Enzyme|enzyme]], the [[wikipedia:Binding_site|binding site]] of a [[wikipedia:Ligand|ligand]], or a [[wikipedia:Receptor_(biochemistry)|receptor]]. |
||
| + | |||
| + | The flexibility of loop sections sometimes allows them to assume more than one local conformation, such as an “open” and “closed” position that either allows or blocks the protein’s function. |
||
| + | |||
| + | ==Loop Classes== |
||
| + | Loops can be loosely classified according to their shapes. For example, loop regions connecting two adjacent sheets (anti-parallel ones) are called "hairpin loops". The shortest Hairpin Loops, just 2-5 amino acids long, are called reverse turns, or [[U-Turn|U-turns]]. Another common shape is the Omega loop. The beginning and end of the loop sequence will be close together, with middle part open, similar to the shape of the Greek letter omega. |
||
| + | [[Category:Protein Structure]] |
||
| + | [[Category:Glossary]] |
||
Latest revision as of 21:34, 26 January 2018
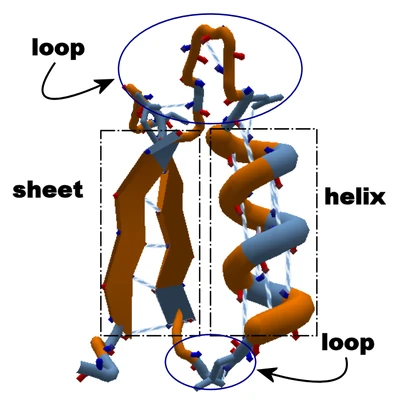
Sections of loop, circled in blue, connect a helix and a sheet, shown in dashed boxes. Blue-and-white spirals show hydrogen bonds. The loop at the top has seven segments, making a relatively long loop.
Loop is one of three types of secondary structure seen in Foldit. Unlike helix and sheet, loop doesn't have a predictable shape.
In Foldit, using one of the "cartoon" view options, sections of loop appear as a thin tube. Loop doesn't have the heavy tube and spiral shape of a helix, or the flat zigzag shape of sheets.
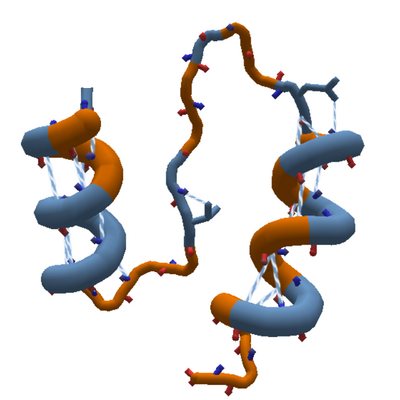
A 12-segment loop connects two helixes. This pattern is found in the natural protein 2FTK, the basis of "Rosetta Decoy 10", seen periodically as a revisiting puzzle. Ideal loops in Foldit are normally much shorter, but real proteins may contain long sections of loop.
Loop doesn't form the regular patterns of hydrogen bonds between backbone atoms that define helix and sheet. The lack of a regular pattern of bonds helps make loop sections more flexible than sheet or helix. In Foldit terms, however, any unbonded atoms represent a missed scoring opportunity, so there's an incentive to keep loop sections short, at least in design puzzles.
In real proteins, there are often long sections of loop that meander between helixes and sheets.
In design puzzles, Foldit scientists have identified loop sections as a problem area. See Ideal Loops for discussion of the new features added to the game to address this issue. With more ideal loops, players are able to design proteins that actually fold up on their own. See design puzzle results for the latest on successful player designs.
The Blueprint tool contains predefined shapes that can be used as a starting point for designing ideal loops. The loop portions of these shapes are short -- only two to five segments. It's possible to design longer loops that satisfy the "ideal loops" condition found in many design puzzles, but it's not necessarily easy.
The Purpose of Loops[]
Helixes and sheets tend to stabilize the core of the protein. Loops are more likely to be found near the surface of the protein. Not surprisingly, loops tend to be rich in hydrophilic sidechains. The hydrophilics in loops make hydrogen bonds with the surrounding water more than with adjacent amino acids, helping make loops more flexible than helices and sheets.
Far from just being “connectors,” loops are often involved in the function of the protein, such as being part of the active site of an enzyme, the binding site of a ligand, or a receptor.
The flexibility of loop sections sometimes allows them to assume more than one local conformation, such as an “open” and “closed” position that either allows or blocks the protein’s function.
Loop Classes[]
Loops can be loosely classified according to their shapes. For example, loop regions connecting two adjacent sheets (anti-parallel ones) are called "hairpin loops". The shortest Hairpin Loops, just 2-5 amino acids long, are called reverse turns, or U-turns. Another common shape is the Omega loop. The beginning and end of the loop sequence will be close together, with middle part open, similar to the shape of the Greek letter omega.