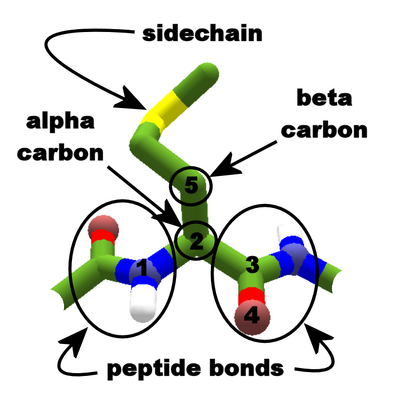
Protein backbone features. Peptide bonds join the carboxyl group of one segment to the amino group of the next. Here, a methionine sidechain is attached to the alpha carbon of the segment in the center. Atom numbers are shown in black. (Stick + Polar H view, EnzDes coloring.)
Protein backbone is what holds a protein together and gives it an overall shape (or tertiary structure).
Compared to RNA and DNA backbone, protein backbone has a relatively simple chemical structure - a nitrogen atom, two carbon atoms, one or two oxygen atoms, and a few hydrogens. (Compare the backbone shown here to RNA backbone, which has over twice as many atoms.)
Protein sidechains receive most of the attention, but understanding the backbone is also important in Foldit. The "cartoon" views in Foldit show the shape of the backbone, but hide most of the chemical structure. Switching to a view like "Stick + Polar H" and using EnzDes or CPK coloring reveals more about the backbone.
Each segment of a protein is the residue of an amino acid. Strong peptide bonds join the segments, forming the backbone. Except for the ends of a protein chain, the backbone of each segment contains the same atoms.
The alpha carbon (atom 2) is the central feature of the backbone. The alpha carbon connects the following parts of the amino acid:
The critical backbone angles named phi and psi are determined relative to the alpha carbon. Phi and psi can be viewed in the Rama map tool in Foldit.
The backbone can form hydrogen bonds at two points: the nitrogen in the amino group (atom 1) can act as a hydrogen bond donor. The oxygen(s) (atom 4, and sometimes atom 5) in the carboxyl group can accept hydrogen bonds. Patterns of hydrogen bonds between these backbone points determine the secondary structure of the protein.
Amino group[]
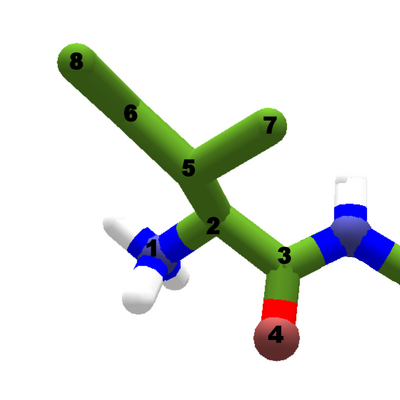
An isoleucine residue as the N terminal of a protein chain. The amino nitrogen at atom 1 has three hydrogens attached.
The backbone of each amino acid starts with the same collection of atoms, but changes occur as peptide bonds are formed. The amino group starts out as a nitrogen atom bonded to three hydrogen atoms. The nitrogen atom is always assigned atom number 1, both Foldit and in science tools like the PDB.
The amino group of the first segment in a chain, known as the N terminal, keeps all three hydrogen atoms. For subsequent segments, the amino group loses two of its hydrogen atoms.
The amino group as seen in Foldit is in its "ionized" or charged form, with three hydrogens, formula NH3-, and a positive charge. The ionic form is what would normally be found in the "aqueous" or water-based solution inside a cell. The amino group can also be shown in it's non-ionic form, with a nitrogen and only two hydrogens, formula NH2-.
Carboxyl group[]
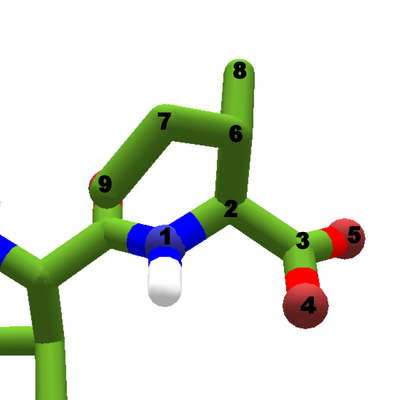
Isoleucine as the C terminal of a protein chain. The carboxyl carbon has two oxygen atoms attached.
Like the amino group, the carboxyl group changes as segments are added to the protein. The carboxyl group starts out as a carbon atom and two oxygen atoms. As the peptide bond forms, the carboxyl loses an oxygen. This change is referred to as a condensation reaction, because the two hydrogens and an oxygen freed up as the peptide bond forms are also combined to form a water molecule (H2O).
Oxygen is a heavy atom, and since it's part of the backbone, the atom numbering for the sidechain changes. For every atom in a protein chain except the last one (the C terminal), the sidechain starts with the beta carbon, atom 5. The C terminal still has two oxygen atoms in its carboxyl group, so its beta carbon is atom 6.
Like the amino group, the carboxyl group is shown in ionized form. So it's formula is -COO, and it has a negative charge. In it's non-ionized form, a carboxyl has the formula -COOH.
Sidechain[]

Isoleucine in the middle of a protein chain. Both the amino and carboxyl groups are part of peptide bonds. The atom numbering is the same as for the N terminal.
The sidechain is what makes each amino acid different. The Amino Acid Gallery shows all sidechains seen in Foldit.
In chemical diagrams, the sidechain may be replaced by the letter R. The wikipedia article "side chain" explains this use of "R" in detail.
Many amino acid sidechains contain nitrogen and oxygen atoms that can act as hydrogen bond donors, acceptors, or both. The Sidechain Bonding Gallery shows the atoms in question, and the Sidechain Bonding Table gives the atom numbers.
Glycine[]
In every amino acid except glycine, the sidechain starts with the beta carbon, atom 5 in every position except the C terminal.

Glycine in the middle of a protein chain. Glycine has no sidechain, and only 7 atoms. (Stick+H view.)
Glycine has no sidechain, just a hydrogen atom filling in for the beta carbon. The lack of a sidechain makes glycine the most flexible amino acid.
Proline[]
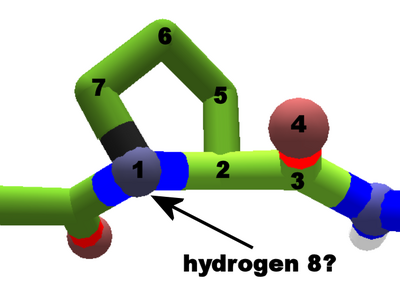
Proline in the middle of a protein chain. Proline's sidechain loops around to the backbone. Foldit doesn't show a hydrogen which should be next to atom 1.
Proline is another exception, with a sidechain which loops around to the backbone. The sidechain starts normally, but ends up bonded to the nitrogen at atom 1.
This unusual structure makes proline relatively inflexible. It's structure also led proline to be classifed as an imino acid at one point in time, but this usage is now considered obsolete.
There should be a hydrogen atom next to the nitrogen at atom 1, but Foldit doesn't show it. The "stick+H" or "line+H" views sometimes show a stray hydrogen nearby, but banding shows it's not the missing hydrogen, which would be atom 8 in a non-terminal segment.
Cutpoints[]
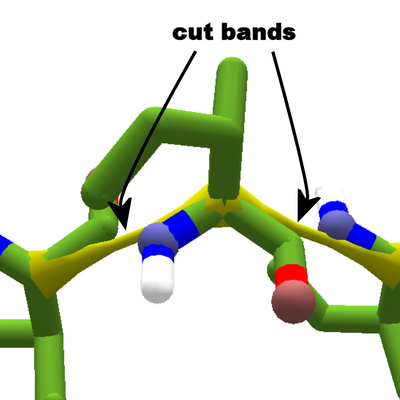
Isoleucine with cutpoints on either side. Yellow cut bands pull toward the alpha carbon.
Foldit allows players to temporarily break peptide bonds using cutpoints.
A cutpoint breaks the bond between the carboxyl carbon of one segment and the amino nitrogen of another. The bond is replaced by a cut band, which is intially yellow. The cut band turns blue if the two segments move too far apart.
Cut bands connect the alpha carbons of the two segments on either side of the cut. So they don't pull on the atoms directly involved the peptide bond.
Cutpoints can be closed when the cut band is yellow by clicking on the cut band. Recipes can open and close cutpoints, but a recipe can't tell if a cutpoint can be closed. Recipes may sometimes leave open cutpoints as a result.