(Adding categories) |
LociOiling (talk | contribs) (Replaced Nature link with Pubmed, converted to wiki markup with light edits throughout.) |
||
| Line 1: | Line 1: | ||
| + | [[File:Koga&Koga.png|thumb|400px|The Koga & Koga paper as seen in PubMed.]] |
||
| + | In 2012, the Baker group published a paper "Principles for designing ideal protein structures” in Nature. This article is usually referred to as "Koga & Koga" in Foldit, after the lead authors. It's the basis of the [[Ideal Loops|ideal loops]] condition found in most recent Foldit [[Design puzzle|design puzzles]], and the patterns found the [[Blueprint|blueprint]] tool. |
||
| + | If you're comfortable with the rather dry language of academic biology, you can read the [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3705962/ complete article on PubMed]. |
||
| − | In 2012, the Baker group published a paper in Nature entitled : "Principles for designing ideal protein structures” : here's the reference if you're comfortable with the rather dry language of academic biology http://www.nature.com/nature/journal/v491/n7423/full/nature11600.html. Based partly on known structures of naturally occurring proteins, and partly on large scale simulations of artificial proteins made by the volunteer contributors of Rosetta@home, the work laid down guidelines as to what kinds of secondary structure patterns would be most promising when designing new proteins. These rules have been explained before in a video by Susume here https://www.youtube.com/watch?v=uXrQ2VWsPJ0: sometimes though its easier to have a written reference for this kind of thing. So here's an attempt to summarize the main features of that paper: if when designing proteins you can follow these guidelines there's a much better chance of that design being interesting (not to mention achieving a higher score). |
||
| + | Koga & Koga describes promising [[Secondary Structure|secondary structure]] patterns for designing new proteins. The patterns are based partly on known structures of naturally occurring proteins, and partly on large-scale simulations of artificial proteins by Rosetta@home and its network of volunteers. |
||
| − | In almost all the following cases it doesn't matter what kind of amino acids are present in any structure element. (exception : hairpin loops in FoldIt are much easier to construct if make the loops residues glycine): the mutate function may change them later. |
||
| + | All of these patterns involve a section of [[sheet]] or [[helix]] connected to another sheet or helix by a short section of [[loop]]. So each pattern involves [[Segment|segments]] (residues) that are immediately adjacent. In natural proteins, sheets are often bonded to other sheets from "distant" parts of the protein, and sometimes loops meander for many segments, but Koga & Koga didn't address these cases. |
||
| − | Here are the structure patterns: |
||
| ⚫ | |||
| ⚫ | |||
| + | The page attempts to summarize the Koga & Koga patterns. If you can use these patterns when designing proteins, there's a much better chance of your design being interesting from a scientific standpoint. The patterns may help in clearing those ideal loops conditions and achieving a higher score. |
||
| ⚫ | |||
| + | The use of these patterns in Foldit is also explained in a [https://www.youtube.com/watch?v=uXrQ2VWsPJ0 video by Susume]. |
||
| ⚫ | |||
| + | Koga & Koga has patterns for sheet - loop - sheet, sheet - loop - helix, and helix - loop - sheet. |
||
| ⚫ | |||
| ⚫ | |||
| + | Koga and Koga didn't include a helix - loop - helix pattern. Fortunately, the Foldit blueprint tool does include samples for helix - loop - helix, along with the other Koga & Koga patterns. |
||
| + | Each of these patterns is discussed in more detail below. The goal is to identify the key features of each pattern and describe how to create the pattern by "hand folding" in Foldit. You can also select similar patterns using the [[Blueprint|blueprint]] tool. Hand folding usually involves using [[Cutpoints|cutpoints]] and the [[Move|move tool]] to get the helixes, sheets, and loops aligned; the blueprint shapes eliminate that step. |
||
| + | With these patterns, it generally doesn't matter which [[Amino Acids|amino acids]] are used at the start. One exception: hairpin loops in Foldit are much easier to construct if you make them [[Glycine|glycine]] to start. The [[Mutate|mutate]] tool can always fine-tune the amino acids later on. |
||
| ⚫ | |||
| + | The segment order of the patterns '''does''' matter. Helix-loop-sheet is not the same as sheet-loop-helix from a folding standpoint. The lower-numbered segments are always listed first in the patterns. |
||
| ⚫ | |||
| + | Several of the patterns involve two adjacent sheets connected by a short section of loop. This is called an "anti-parallel" arrangment, and its very common in natural proteins. |
||
| ⚫ | |||
| ⚫ | In the case of this particular secondary structure sequence |
||
| − | + | The method for determining "left" and "right" is discussed for the sheet - 2 segment loop - sheet pattern. |
|
| + | <br clear="all"/> |
||
| ⚫ | |||
| + | [[File:E-L2_E.jpg|thumb|400px|Sheet connected to sheet by two loop segments.]] |
||
| + | This pattern is seen in many natural proteins. Here's a strategy to make this pattern manually in Foldit. |
||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| + | To see the sidechains, turn on "show sidechains (all)" in the Foldit [[View Options|view options]]. You'll also want to turn on "show bonds (sheet)" to show the blue-and-white spirals that indicate the sheets are properly aligned. |
||
| ⚫ | |||
| ⚫ | In this example, sheet 1 has segments 55 to 59. Segments 60 and 61 constitute the loop. The second sheet has segments 62 to 66. Note that the sidechain of segment 59 ([[arginine]]) points into the page, indicating that the second sheet goes to the left. Also note that one of the loop segments (61) shows no sidechain, which means it's a [[Glycine|glycine]]. |
||
| ⚫ | |||
| ⚫ | In the case of this particular secondary structure sequence, the preference for going left might be considered an absolute rule rather than a guideline. In thousands of simulated cases where this pattern occurred, and many thousands more in naturally occurring proteins, it looks from the Koga & Koga paper as if there was not a single case of the second sheet being to the right when the sheets are joined by a 2-segment loop. |
||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
It's 50/50 whether the second sheet goes right or left in both natural and artificial proteins, so you don't have to worry about it unduly. |
It's 50/50 whether the second sheet goes right or left in both natural and artificial proteins, so you don't have to worry about it unduly. |
||
| − | + | This pattern occurs about twice as frequently in natural proteins as the 3-segment loop case, but is still relatively uncommon relative to the 2-segment loop case. |
|
| ⚫ | |||
| + | [[File:E-5L-E.jpg|thumb|400px|Sheet connected to sheet by five loop segments.]] |
||
| ⚫ | |||
| − | + | #Then the second sheet should be to the RIGHT of the first sheet. |
|
| − | |||
| ⚫ | |||
| − | |||
| − | 2) Then the second sheet should be to the RIGHT of the first sheet. |
||
| − | [[File:E-5L-E.jpg|thumb|left|400px]] |
||
| − | |||
| − | |||
| − | |||
| ⚫ | |||
| − | |||
| − | |||
| − | |||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
It's also much more common in naturally occurring proteins than the 3 and 4 loop cases but still not as frequent as the 2-loop case. |
It's also much more common in naturally occurring proteins than the 3 and 4 loop cases but still not as frequent as the 2-loop case. |
||
| − | + | ==sheet - loop - helix== |
|
| ⚫ | |||
| − | |||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| + | [[File:E-2L-H.jpg|thumb|400px|Sheet connected to helix by two loop segments.]] |
||
Here, the preference is for the helix to go behind the sheet and slant to the left as shown below. |
Here, the preference is for the helix to go behind the sheet and slant to the left as shown below. |
||
| ⚫ | |||
| ⚫ | |||
| − | [[File:E-2L-H.jpg|thumb|left|400px]] |
||
| ⚫ | |||
| ⚫ | |||
| − | |||
| − | |||
| − | |||
| ⚫ | |||
| − | |||
| − | |||
| − | |||
| ⚫ | |||
| − | |||
| ⚫ | |||
| + | <br clear="all"/> |
||
| + | ===sheet - 3 segment loop - helix=== |
||
| + | [[File:E_3L-H.jpg|thumb|400px|Sheet connected to helix by three loop segments.]] |
||
Here, the preference is for the helix to go in front of the sheet and slant to the right as shown below. |
Here, the preference is for the helix to go in front of the sheet and slant to the right as shown below. |
||
| ⚫ | |||
| ⚫ | |||
| − | [[File:E_3L-H.jpg|thumb|left|400px]] |
||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| ⚫ | |||
| − | |||
| − | |||
| − | |||
| ⚫ | |||
| − | |||
| ⚫ | |||
| − | |||
| ⚫ | |||
| − | [[File:H-L-E.jpg|thumb|left|400px]] |
||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| ⚫ | |||
| + | <br clear="all"/> |
||
| − | The paper doesn't mention these: not sure why they wouldn't be worthy of a mention. |
||
| + | ==helix - loop - sheet== |
||
| ⚫ | |||
| ⚫ | |||
[[Category:Structure]] |
[[Category:Structure]] |
||
[[Category:Protein Structure]] |
[[Category:Protein Structure]] |
||
Revision as of 23:18, 13 January 2018

The Koga & Koga paper as seen in PubMed.
In 2012, the Baker group published a paper "Principles for designing ideal protein structures” in Nature. This article is usually referred to as "Koga & Koga" in Foldit, after the lead authors. It's the basis of the ideal loops condition found in most recent Foldit design puzzles, and the patterns found the blueprint tool.
If you're comfortable with the rather dry language of academic biology, you can read the complete article on PubMed.
Koga & Koga describes promising secondary structure patterns for designing new proteins. The patterns are based partly on known structures of naturally occurring proteins, and partly on large-scale simulations of artificial proteins by Rosetta@home and its network of volunteers.
All of these patterns involve a section of sheet or helix connected to another sheet or helix by a short section of loop. So each pattern involves segments (residues) that are immediately adjacent. In natural proteins, sheets are often bonded to other sheets from "distant" parts of the protein, and sometimes loops meander for many segments, but Koga & Koga didn't address these cases.
The page attempts to summarize the Koga & Koga patterns. If you can use these patterns when designing proteins, there's a much better chance of your design being interesting from a scientific standpoint. The patterns may help in clearing those ideal loops conditions and achieving a higher score.
The use of these patterns in Foldit is also explained in a video by Susume.
Koga & Koga has patterns for sheet - loop - sheet, sheet - loop - helix, and helix - loop - sheet.
Koga and Koga didn't include a helix - loop - helix pattern. Fortunately, the Foldit blueprint tool does include samples for helix - loop - helix, along with the other Koga & Koga patterns.
Each of these patterns is discussed in more detail below. The goal is to identify the key features of each pattern and describe how to create the pattern by "hand folding" in Foldit. You can also select similar patterns using the blueprint tool. Hand folding usually involves using cutpoints and the move tool to get the helixes, sheets, and loops aligned; the blueprint shapes eliminate that step.
With these patterns, it generally doesn't matter which amino acids are used at the start. One exception: hairpin loops in Foldit are much easier to construct if you make them glycine to start. The mutate tool can always fine-tune the amino acids later on.
The segment order of the patterns does matter. Helix-loop-sheet is not the same as sheet-loop-helix from a folding standpoint. The lower-numbered segments are always listed first in the patterns.
sheet - loop - sheet
Several of the patterns involve two adjacent sheets connected by a short section of loop. This is called an "anti-parallel" arrangment, and its very common in natural proteins.
The key here is the number of segments in the loop, which determines whether the second sheet goes to the left or right of the first sheet.
The method for determining "left" and "right" is discussed for the sheet - 2 segment loop - sheet pattern.
sheet - 2 segment loop - sheet

Sheet connected to sheet by two loop segments.
This pattern is seen in many natural proteins. Here's a strategy to make this pattern manually in Foldit.
- Arrange the first sheet (the one with lower segment numbers) so that the sidechain of the last segment of the sheet points into the screen.
- Then the second sheet should be to the LEFT of the first sheet.
- When constructing this hairpin turn, life is much easier if you mutate the two loop segments to glycine. Also, put a cutpoint in the middle of the loop and wiggle it to get a reasonable shape.
To see the sidechains, turn on "show sidechains (all)" in the Foldit view options. You'll also want to turn on "show bonds (sheet)" to show the blue-and-white spirals that indicate the sheets are properly aligned.
In this example, sheet 1 has segments 55 to 59. Segments 60 and 61 constitute the loop. The second sheet has segments 62 to 66. Note that the sidechain of segment 59 (arginine) points into the page, indicating that the second sheet goes to the left. Also note that one of the loop segments (61) shows no sidechain, which means it's a glycine.
In the case of this particular secondary structure sequence, the preference for going left might be considered an absolute rule rather than a guideline. In thousands of simulated cases where this pattern occurred, and many thousands more in naturally occurring proteins, it looks from the Koga & Koga paper as if there was not a single case of the second sheet being to the right when the sheets are joined by a 2-segment loop.
sheet - 3 segment loop - sheet
This pattern is very similar to the previous case. The second sheet has a strong preference to go to to the left.
- Arrange the first sheet (the one with lower segment numbers) so that the sidechain of the last segment of the sheet points into the screen.
- Then the second sheet should, as previously, be to the LEFT of the first sheet.
- The loop here isn't quite as strained as in the 2-segment loop case above so glycines in the loop aren't a necessity during construction. Mutate may still end up putting them there though.
This preference is followed about 85% of the time (based on eyeballing the histograms in the paper), both in naturally occurring proteins and in designed ones.
This pattern occurs much less frequently in natural proteins than does the 2-segment loop case (maybe 10-15% as common).
sheet - 4 segment loop - sheet
It's 50/50 whether the second sheet goes right or left in both natural and artificial proteins, so you don't have to worry about it unduly.
This pattern occurs about twice as frequently in natural proteins as the 3-segment loop case, but is still relatively uncommon relative to the 2-segment loop case.
sheet - 5 segment loop - sheet

Sheet connected to sheet by five loop segments.
- Arrange the first sheet (the one with lower segment numbers) so that the sidechain of the last segment of the sheet points into the screen.
- Then the second sheet should be to the RIGHT of the first sheet.
In this example, sheet 1 has segments 4 to 8. Segments 9 to 13 make up the loop, and the second sheet has segments 14 to 18. Note that the sidechain of segment 8 (serine: barely visible) points into the page, indicating that the second sheet goes to the right.
This pattern is followed about 70% of the time in designed proteins and about 95% of the time in naturally occurring ones.
It's also much more common in naturally occurring proteins than the 3 and 4 loop cases but still not as frequent as the 2-loop case.
sheet - loop - helix
There are two preferred orientations for this setup. In both of them the helix is offset diagonally from the sheet: in one case it is in front of the sheet and slants to the right. In the other case, the helix goes behind the sheet and slants to the left.
sheet - 2 segment loop - helix

Sheet connected to helix by two loop segments.
Here, the preference is for the helix to go behind the sheet and slant to the left as shown below.
To avoid visual clutter, only the last sidechain (segment 9) of the sheet is shown: pointing into the page as usual to provide a defined orientation. The loop (segments 10 to 11) and the start of the helix (segment 12) are also shown.
When the loop is two segments in length, this orientation is favoured over the one below by about 10-1.
It's actually quite hard in Foldit to achieve this geometry without the helix and sheet getting too close: furthermore, the distinction between a loop and a helix isn't all that clear.
sheet - 3 segment loop - helix
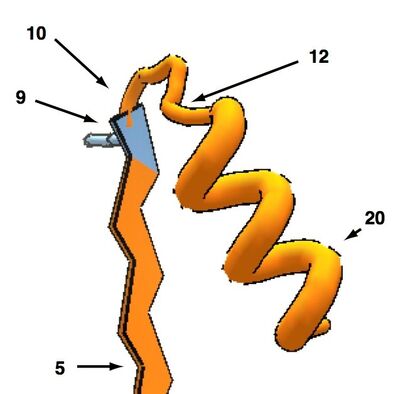
Sheet connected to helix by three loop segments.
Here, the preference is for the helix to go in front of the sheet and slant to the right as shown below.
To avoid visual clutter, only the last sidechain (segment 9) of the sheet is shown: pointing into the page as usual to provide a defined orientation. The loop (segments 10 to 12) and the start of the helix ( segment 12) are alos shown.
This orientation is favoured over the two-loop arrangment in both natural and designed proteins but in neither case is the preference overwhelming: it's about 2-1 in artificial proteins and 1.5 to 1 in naturally occurring ones.
helix - loop - sheet

Helix connected to sheet.
Irrespective of the size of the loop (bit peculiar that), the orientation shown here is preferred. Again the helix is at an angle to the sheet, the first segment of which points into the plane.