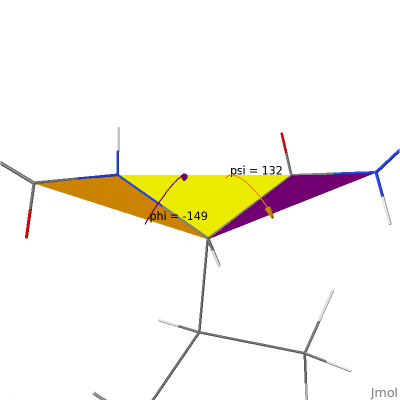
Animation of phi and psi. The yellow triangle is the reference plane. Phi is measured between the orange and yellow triangles. Psi is measured between the purple and the yellow triangles. In this line view with hydrogens displayed, the sidechain of an isoleucine points down from the point of the yellow triangle. Generated from Psi and Phi angles at protopedia.org
Backbone angles in a protein are dihedral angles, which are the angles between two intersecting planes. They're also referred to as torsion angles, referring to how part of the backbone rotates or twists twists with regard to another part.
The most important backbone angles are phi (φ) and psi (ψ). These angles are plotted on the Rama map. Phi is the horizontal x-axis, psi is the vertical y-axis of the Rama map.
Both phi and psi involve the alpha carbon (or Cα), the atom that connects the sidechain, the amino group, and the carboxyl group, making it the center of a segment's backbone.
The other backbone angle is omega (ω), which involves the peptide bond between two segments. The peptide bond is expected to be nearly "planar" or flat, so this angle is considered less interesting. Unlike phi and psi, which are shown in the Rama map, Foldit doesn't have a direct display of omega angles.
Measuring phi and psi[]
Planes are determined by three points. The reference plane for both phi and psi contains three atoms: the nitrogen in the amino group, the alpha carbon, and the carbon in the carboxyl group. (The carboxyl carbon is sometimes indicated as C' or "C prime".)
The plane for phi contains the carboxyl carbon from the previous segment, plus the amino nitrogen and the alpha carbon from the segment in question. Phi is the angle between this plane and the reference plane.
The plane for psi contains the amino nitrogen from the next segment, plus carboxyl carbon and the alpha carbon from the segment in question. Psi is the angle between this plane and the reference plane.
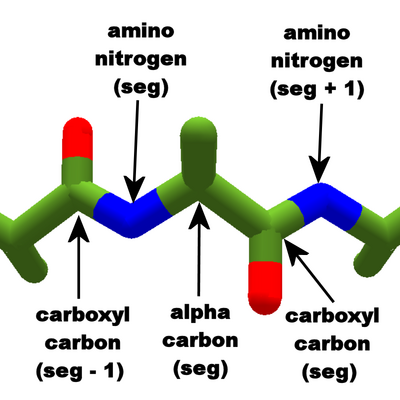
The component atoms that determine phi and psi. Phi involves the four atoms on the left; psi involves the four atoms on the right.
The planes for phi and psi both share two atoms with the reference plane. This allows phi and psi (and dihedral angles in general) to be specified by just four points.
Phi and psi involve atoms in the segment for which the angles are being measured (seg), the previous segment (seg - 1), and the next segment (seg + 1).
For phi, the four points are the locations of the carboxyl carbon (seg - 1), the amino nitrogen (seg), the alpha carbon (seg), and the carboxyl carbon (seg).
For psi, the four points are the locations of the amino nitrogen (seg + 1), the carboxyl carbon (seg), the alpha carbon (seg), and the amino nitrogen (seg).
Measuring omega[]
Omega is again a dihedral angle, involving four atoms, just like phi and psi.
For omega, the four atoms are the alpha carbon (seg - 1), the carboxyl carbon (seg - 1), the amino nitrogen (seg), and the alpha carbon (seg).
Usually, omega is around 180°, which is referred to as the trans case. More rarely, omega is around 0°, which is called the cis case. The trans configuration means the sidechains of successive segments end up on opposite sides of the protein's backbone. This is how proteins appear in the extended chain at the start of Foldit design and de-novo puzzles.
The most common cis configuration occur when proline follows another amino acid, sometimes referred to as an "X-P" bond.
What rotates?[]
Phi, psi, and omega are known torsion angles. Foldit allows phi and psi to be adjusted directly in the Rama map, but it can be difficult to predict the impact on the protein.
In effect, phi rotates the two segments (seg - 1 and seg) around the bond between the amino nitrogen and alpha carbon of the segment in question (seg). As seen in this animation, demonstrates the rotation, showing the previous segment's carboxyl group spinning around the N-Cα axis of the segment in question.
Similarly, psi rotates two segments (seg and seg + 1) around the bond between the alpha carbon and the carboxyl carbon (Cα-C') of the segment in question. Again, an animation demonstrates the rotation, showing the carboxyl group and the next segment's amino group spinning around.
Both these animations show one side spinning while the other remains still, but of course that depends on your point of view.
Omega angle rotation doesn't have an animated image available, but it's easy to visualize. If you were sighting along the backbone, the sidechains of two trans segments would start out at the twelve o'clock and six o'clock positions. Changing omega means the sidechains rotate relative to each other. Keeping the 12 o'clock sidechain still, the six o'clock sidechain might move toward the five or the seven o'clock position.
Another way of looking at this type of rotation is to consider the points. Changing a dihedral angle means changing the distance between the first and last of the four points that specify the dihedral. The distances between the other pairs of points don't change.
So, changing the value of phi changes the distance between the carboxyl carbons of two segments. Changing the value of psi change the distance of the amino nitrogens of two segments. And changing omega changes the distance between two alpha carbons.
The distances between other atoms of course also change as phi, psi, or omega changes, but these other atoms aren't involved in specifying the dihedral.
Rotation in Foldit[]
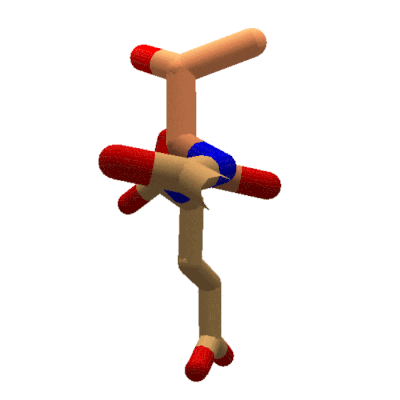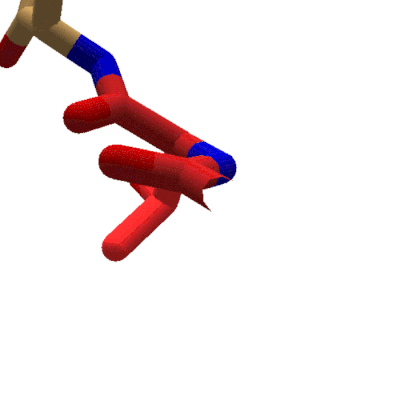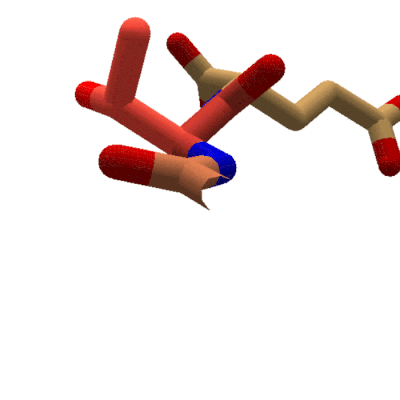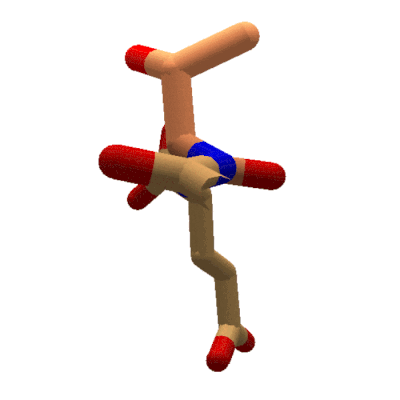
Animation showing the effect of changing phi in Foldit. The sidechain for the tyrosine segment moves while the carboxyl oxygen for the previous segment stays put. The protein takes on red coloring as clashes occur during rotation. (Stick view, ligand-specific coloring.
The animations referenced above show the sidechain of a segment holding steady as the segment's phi and psi values change.
In Foldit, adjusting phi and psi in the Rama map has a different effect on the display. Changing a segment's phi value causes the segment's sidechain to rotate with respect to the lower-numbered segments of the protein. Looking down the backbone toward the higher-numbered segments (toward the C terminal), the backbone for the previous segment doesn't move as phi changes. Instead, the sidechain and backbone for the selected segment (the segment for which the phi value is changing) rotate. All the remaining segments in the protein rotate as well.
Changing psi has a similar effect. Once again looking toward the C terminal, as psi changes, the sidechain for the selected segment doesn't move, but it's carboxyl group and all higher-numbered segments rotate.